产品简介
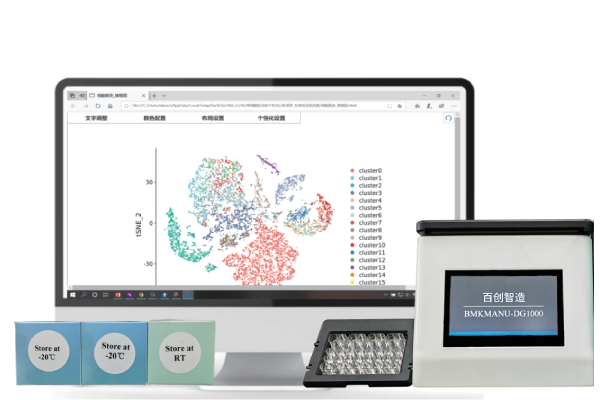
百创DG1000优势

轻巧便携

快速运行

上样灵活
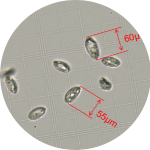
更大细胞直径兼容
技术流程
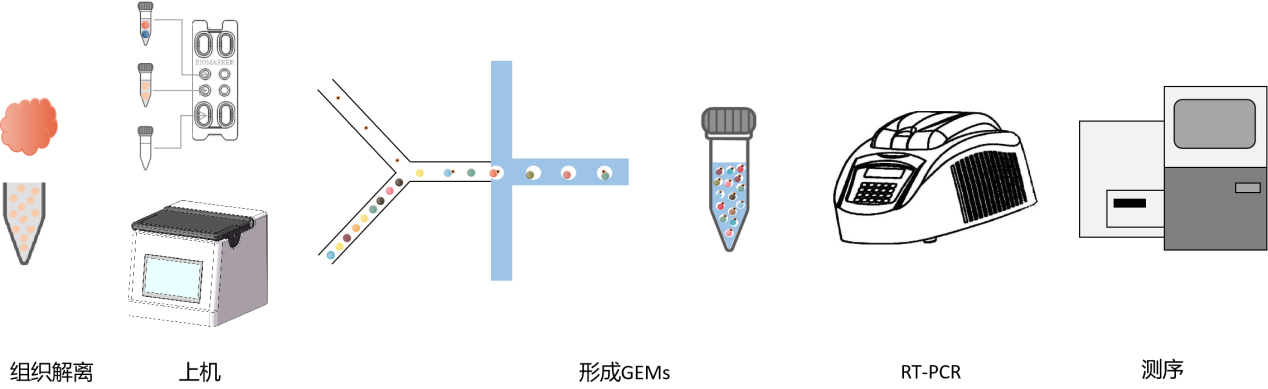
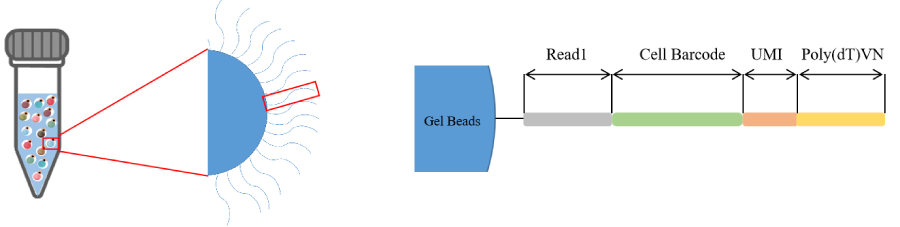
捕获探针
外包科研服务与自己操作区别
| 单细胞测序流程 | 外包科研服务 | 自己操作 |
| 实验周期 | 〜10d | 1〜2d |
| 分析周期 | 〜10d | 1〜2d |
| 是否需要运输 | 运输时间需1〜2d,可能导致细胞活性较低 | 不需运输,现场解离,细胞活性高 |
周期短
保证细胞活性
节约成本
实测数据
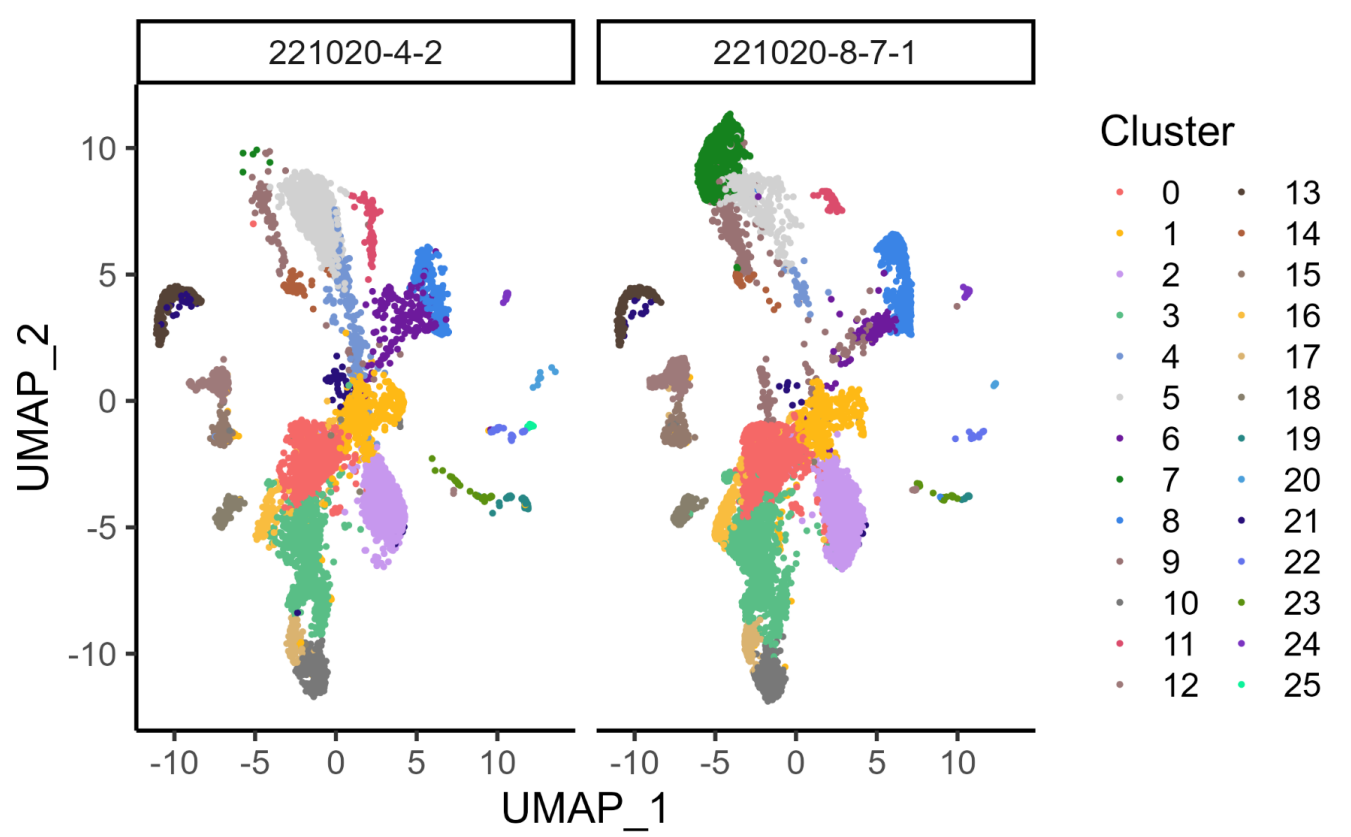
1、测序reads统计
测序reads统计如下表:
表1 测序数据统计
|
Type |
Values 221020-4-2 |
Values 21020-8-7-1 |
|
Number of Reads |
399,967,594 |
378,584,610 |
|
Valid Barcodes |
339,047,468 |
323,897,836 |
|
Valid UMIs |
395,738,572 |
374,651,123 |
注:
Number of Reads:reads总数;
Valid Barcodes:包含有效Barcode的reads数;
Valid UMIs:包含有效UMIs的reads数。
2、数据比对
比对结果统计如下表:
表2 比对结果统计
|
Type |
Values 221020-4-2 |
Values 21020-8-7-1 |
|
Reads Mapped to Genome |
96.01% |
95.52% |
|
Reads Mapped Confidently to Genome |
80.77% |
81.7% |
|
Reads Mapped Confidently to Intergenic Regions |
1.69% |
2.58% |
|
Reads Mapped Confidently to Intronic Regions |
15.62% |
29.15% |
|
Reads Mapped Confidently to Exonic Regions |
63.44% |
49.96% |
|
Reads Mapped Confidently to Transcriptome |
71.64% |
71.57% |
注:
Reads Mapped to Genomes:比对到参考基因组上的Reads在总Reads中占的比例;
Reads Mapped Confidently to Genome:比对到参考基因组并得到转录本GTF信息支持的Reads在总Reads中占的比例;
Reads Mapped Confidently to Intergenic Regions:比对到基因间区域的Reads在总Reads中占的比例;
Reads Mapped Confidently to Intronic Regions:比对到内含子区域的Reads在总Reads中占的比例;
Reads Mapped Confidently to Exonic Regions:比对到外显子区域的Reads在总Reads中占的比例;
Reads Mapped Confidently to Transcriptome:比对到已知参考转录本的Reads在总Reads中占的比例。
3、分析统计
Cell分析统计如下:
表3 Cell统计
|
Type |
Values 221020-4-2 |
Values 21020-8-7-1 |
|
Estimated Number of Cells |
7,104 |
11,012 |
|
Median UMI Counts per Cell |
7,261 |
6,434 |
|
Median Genes per Cell |
2,624 |
2,678.5 |
|
Total Genes Detected |
37,416 |
41,208 |
|
Sequencing Saturation |
73.22% |
65.99% |
|
Fraction Reads in Cells |
77.39% |
83.64% |
注:
Estimated Number of Cells:检测到的细胞数;
Median UMI Counts per Cell:每个cell的UMI中位数;
Median Genes per Cell:每个cell中基因的中位数;
Total Genes Detected:基因总数;
Sequencing Saturation:测序饱和度;
Fraction Reads in Cells:过滤后细胞reads数占总reads数比例。
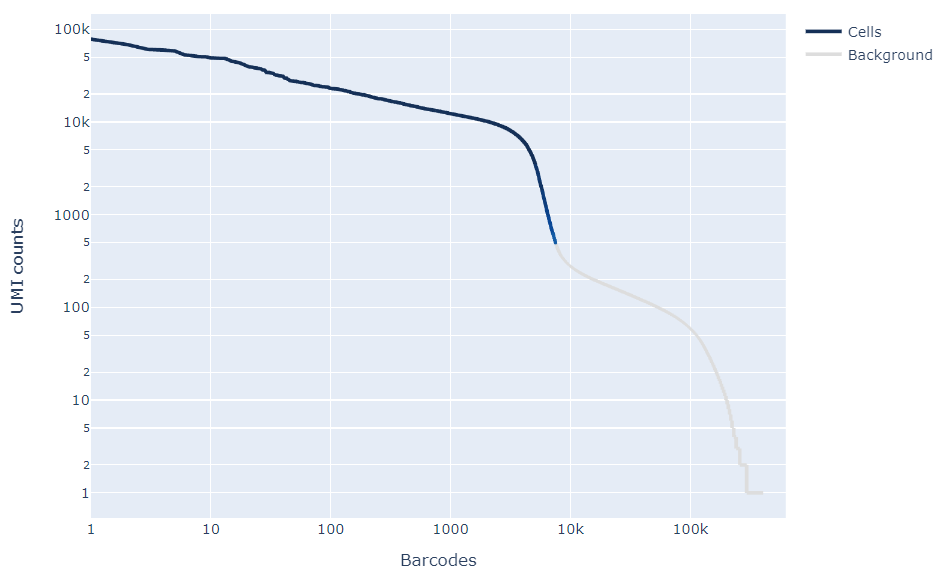
图2 221020-4-2 Barcode rank统计图
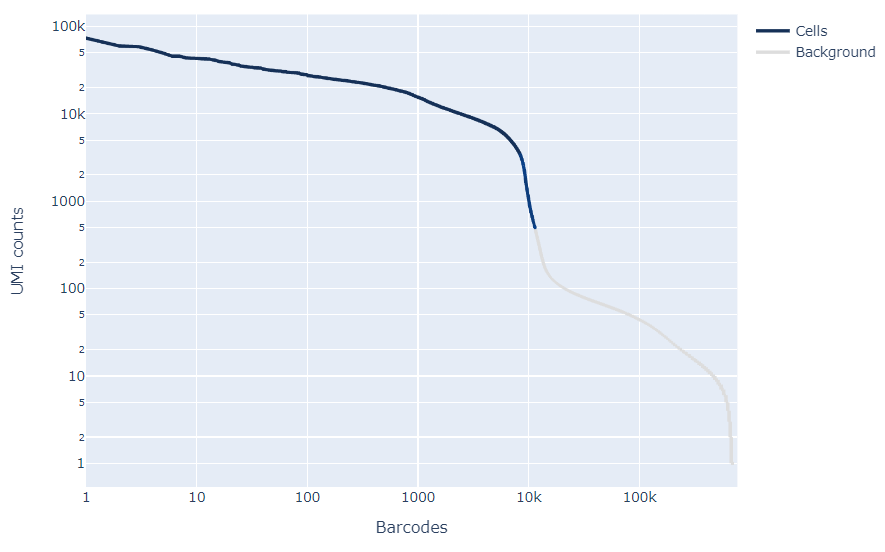
图2 21020-8-7-1Barcode rank统计图
