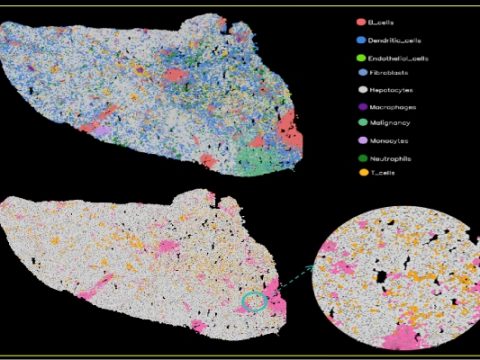
某动物皮肤单细胞Demo数据

BSCMatrix v1.9环境搭建及使用
2022年7月15日
S1000 Seurat分析流程
2022年8月3日
测序reads统计
测序reads统计如下表:
| Type | Values |
| Number of Reads | 187088582 |
| Valid Barcodes | 162097865 |
| Valid UMIs | 175241433 |
注:
Number of Reads:reads总数;
Valid Barcodes:包含有效Barcode的reads数 ;
Valid UMIs:包含有效UMIs的reads数。
数据比对
比对结果统计如下表:
| Type | Values |
| Reads Mapped to Genome | 89.80% |
| Reads Mapped Confidently to Genome | 72.68% |
| Reads Mapped Confidently to Intergenic Regions | 5.79% |
| Reads Mapped Confidently to Intronic Regions | 6.20% |
| Reads Mapped Confidently to Exonic Regions | 60.68% |
| Reads Mapped Confidently to Transcriptome | 54.81% |
注:
Reads Mapped to Genomes:比对到参考基因组上的Reads在总Reads中占的比例;
Reads Mapped Confidently to Genome:比对到参考基因组并得到转录本GTF信息支持的Reads在总Reads中占的比例;
Reads Mapped Confidently to Intergenic Regions:比对到基因间区域的Reads在总Reads中占的比例;
Reads Mapped Confidently to Intronic Regions:比对到内含子区域的Reads在总Reads中占的比例;
Reads Mapped Confidently to Exonic Regions:比对到外显子区域的Reads在总Reads中占的比例;
Reads Mapped Confidently to Transcriptome:比对到已知参考转录本的Reads在总Reads中占的比例。
分析统计
Cell分析统计如下:
| Type | Values |
| Estimated Number of Cells | 3376 |
| Median UMI Counts per Cell | 5504.5 |
| Median Genes per Cell | 2069.5 |
| Total Genes Detected | 26262 |
| Sequencing Saturation | 76.38% |
| Fraction Reads in Cells | 87.72% |
注:
Estimated Number of Cells:检测到的细胞数;
Median UMI Counts per Cell:每个cell的UMI中位数;
Median Genes per Cell:每个cell中基因的中位数;
Total Genes Detected:基因总数;
Sequencing Saturation:测序饱和度;
Fraction Reads in Cells:过滤后细胞reads数占总reads数比例。
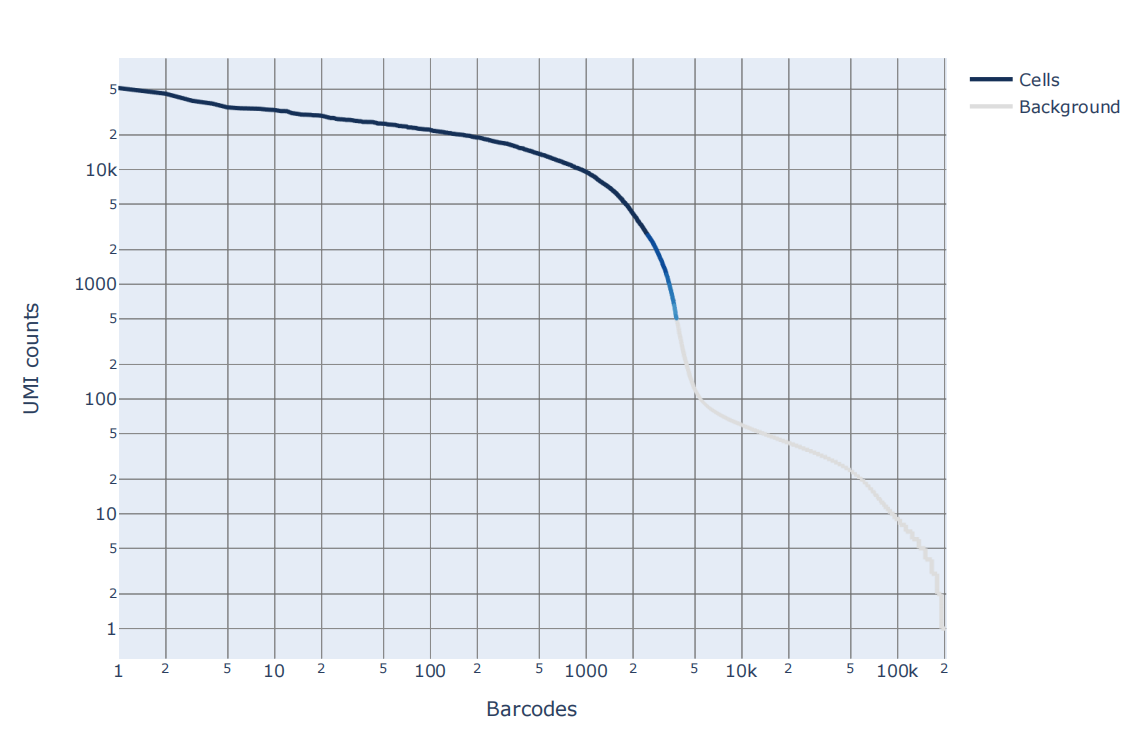
Barcode rank统计图